Concepts of the MoMaLab
Two contact manipulation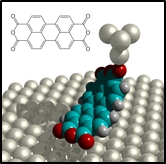 In two-contact manipulation the molecule is simultaneously connected to both the surface and the tip. It is a promising approach in the context of molecular fabrication because the SPM tip and the surface act together like two robotic fingers that manipulate a molecule which is bridging them. If the tip is displaced relative to the sample, this gives access to internal molecular degrees of freedom. Moreover, two-contact manipulation can be carried out as a completely continuous process, pretty much like a robot would handle a workpiece. A current limitation of two-contact manipulation is the fact that one of the two contacts, the surface, is static and needs to play the role of an actuator and the work bench at the same time.
In two-contact manipulation the molecule is simultaneously connected to both the surface and the tip. It is a promising approach in the context of molecular fabrication because the SPM tip and the surface act together like two robotic fingers that manipulate a molecule which is bridging them. If the tip is displaced relative to the sample, this gives access to internal molecular degrees of freedom. Moreover, two-contact manipulation can be carried out as a completely continuous process, pretty much like a robot would handle a workpiece. A current limitation of two-contact manipulation is the fact that one of the two contacts, the surface, is static and needs to play the role of an actuator and the work bench at the same time.
Manipulation in 3D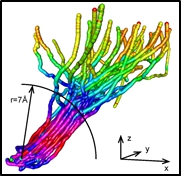 Today’s SPM-based manipulation events are typically of an abrupt and often stochastic nature. This is unavoidable for small and compact entities in which the center-of-mass coordinate is the only relevant descriptor and for which the manipulation thus essentially reduces to either dragging, pushing or a pick up-transfer-drop sequence. In these manipulations, the manipulated object is either attached to the tip or to the sample surface. The situation is entirely different for complex objects with many accessible degrees of freedom and anisotropic shapes. These can bind to both tip and sample simultaneously in many orientations and different internal configurations. Therefore, additional descriptors come into play. While this complicates the manipulation process considerably, it does create the possibility to actively control the orientation and configuration of the manipulated object. Evidently, this is precisely what is needed for directed molecular fabrication, if, e.g., an additional molecule is to be added to a molecular nanostructure in a well-defined position and orientation.
Today’s SPM-based manipulation events are typically of an abrupt and often stochastic nature. This is unavoidable for small and compact entities in which the center-of-mass coordinate is the only relevant descriptor and for which the manipulation thus essentially reduces to either dragging, pushing or a pick up-transfer-drop sequence. In these manipulations, the manipulated object is either attached to the tip or to the sample surface. The situation is entirely different for complex objects with many accessible degrees of freedom and anisotropic shapes. These can bind to both tip and sample simultaneously in many orientations and different internal configurations. Therefore, additional descriptors come into play. While this complicates the manipulation process considerably, it does create the possibility to actively control the orientation and configuration of the manipulated object. Evidently, this is precisely what is needed for directed molecular fabrication, if, e.g., an additional molecule is to be added to a molecular nanostructure in a well-defined position and orientation.
Real-time simulation Simulations of the manipulation process are required to gain a more detailed understanding of the molecular manipulation process and the molecular conformations. Our ultimate goal in terms of speed is a simulation that runs parallel to a manipulation experiment, providing a real-time analysis of measured data. Currently only simulations on the molecular-mechanics (MM) level are capable of providing such a speed. We develop accurate force fields by parameterizing them to fit experimental data and ab-initio simulations. We do that specifically for the molecule that we want to simulate, which guarantees the highest degree of accuracy possible with a MM approach. We use our simulations two solve the inverse problem related to insufficient measurement data and reconstruct the adsorption potential of molecules on the surface.
more..
Simulations of the manipulation process are required to gain a more detailed understanding of the molecular manipulation process and the molecular conformations. Our ultimate goal in terms of speed is a simulation that runs parallel to a manipulation experiment, providing a real-time analysis of measured data. Currently only simulations on the molecular-mechanics (MM) level are capable of providing such a speed. We develop accurate force fields by parameterizing them to fit experimental data and ab-initio simulations. We do that specifically for the molecule that we want to simulate, which guarantees the highest degree of accuracy possible with a MM approach. We use our simulations two solve the inverse problem related to insufficient measurement data and reconstruct the adsorption potential of molecules on the surface.
more..
Force (gradient) measurement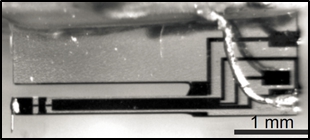 We use a low-temperature ultra-high vacuum combined non-contact atomic force / scanning tunneling microscope (NC-AFM/STM) which uses a qPlus type tuning fork sensor. With tip oscillation amplitudes as low as 0.2 nm, we can precisely measure the change in the tuning fork resonance frequency. This change in frequency is proportional to the force gradient acting on the SPM tip. In this way, we have a direct access to a mechanical property of the tip-molecule-surface junction. This is beneficial since the relation between junction stiffness and molecular conformation is much clearer and easier to simulate than the relation between said conformation and the conductance of the junction which can be measured as well with our setup, but also with STM-only setups.
We use a low-temperature ultra-high vacuum combined non-contact atomic force / scanning tunneling microscope (NC-AFM/STM) which uses a qPlus type tuning fork sensor. With tip oscillation amplitudes as low as 0.2 nm, we can precisely measure the change in the tuning fork resonance frequency. This change in frequency is proportional to the force gradient acting on the SPM tip. In this way, we have a direct access to a mechanical property of the tip-molecule-surface junction. This is beneficial since the relation between junction stiffness and molecular conformation is much clearer and easier to simulate than the relation between said conformation and the conductance of the junction which can be measured as well with our setup, but also with STM-only setups.
Enabling full conformational control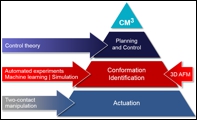 The challenge to enable controlled mechanical manipulation of molecules is the challenge to overcome the complexity gap. This gap has two components. On the one side it is a control gap which opens between a molecule with many degrees of freedom and an actuator, the SPM tip, which has essentially only three degrees of freedom. The second aspect of the complexity gap is the information gap. This is a gap between many degrees of freedom of the molecule and the few measurement channels available from the SPM.
The challenge to enable controlled mechanical manipulation of molecules is the challenge to overcome the complexity gap. This gap has two components. On the one side it is a control gap which opens between a molecule with many degrees of freedom and an actuator, the SPM tip, which has essentially only three degrees of freedom. The second aspect of the complexity gap is the information gap. This is a gap between many degrees of freedom of the molecule and the few measurement channels available from the SPM.
The control gap cannot be overcome fully since it is impossible to control the position of each and every atom in a molecule independently and simultaneously. Our approach therefore focuses on increasing the degree of control possible with only a single actuator. This can for example be achieved by investigating which structural properties of a molecule help to make it more controllable with a single actuator. Another possibility is the engineering of nanostructures on the surface which guide the molecule into a desire to conformation while it is manipulated. The information gap on the other hand results from the imbalance between many degrees of freedom of the molecule and only few measurement channels of the SPM. We can overcome this imbalance by acquiring large amounts of data while manipulating the molecule a small region around its current conformation. As long as this manipulation is fully reversible, the conformation is essentially not changed. We will combine advanced molecular mechanics simulations and machine learning to analyze this data and uniquely assign molecular conformations to the measured data. In this way we will finally realize the dream of being able to handle a single molecule like a macroscopic building block including the possibility to “see” it while it is handled and manipulated and finally the dream of being able to assemble supramolecular structures from these molecules by controlled manipulation. Machine learning We can overcome the information gap discussed above by acquiring large amounts of data while manipulating the molecule a small region around its current conformation. As long as this manipulation is fully reversible, the conformation is essentially not changed. We will implement machine learning solutions (such as (un)supervised learning, kernel ridge regression, support vector machines etc.) for the various data analysis tasks required in this context.
We can overcome the information gap discussed above by acquiring large amounts of data while manipulating the molecule a small region around its current conformation. As long as this manipulation is fully reversible, the conformation is essentially not changed. We will implement machine learning solutions (such as (un)supervised learning, kernel ridge regression, support vector machines etc.) for the various data analysis tasks required in this context.
more.. Control theory Controlled molecular manipulation means knowing the transient conformations and positions of the molecule during manipulation and using this information to reach in a controlled way a target state.
Controlled molecular manipulation means knowing the transient conformations and positions of the molecule during manipulation and using this information to reach in a controlled way a target state.
more.. |
▶January 19th, 2023 MomaLab is part of the Orbital Cinema project!▶September 8th, 2022 Warmest congratulations to Taner Esat for winning the Gerhard Ertl Young Investigator Award for his outstanding work in surface science, particularly metastable standing molecules.▶April 7th, 2022 Want to make your own standing molecule? "Design Principles for Metastable Standing Molecules" Read at J. Phys. Chem. C▶October 10th, 2021 We finally knocked it down: "The stabilization potential of a standing molecule" Read at Science Advances Press release▶October 22nd, 2020 Introducing machine learning to the nanoscale: "Autonomous robotic nanofabrication with reinforcement learning" Read at Science Advances Press release▶July 2020 Our Helmholtz-AI project MomoNano (together with HZB and TU-Berlin) successfully competed for a three year funding by the Helmholtz Association.▶September 2nd, 2020 "The theory of scanning quantum dot microscopy" Read at J Phys. Cond. Mat. |